Explorating the openFDA API part 1 : gathering data
This is a series of posts that explore how to gather, clean, analyze, and generate insights from the data. In this post, I’d like to test out the open FDA API and understand a bit better the data structure behind the API call results, and how we can transform highly nested json into something more familiar, such as a pandas data frame.
Table of Content
- About the open FDA project
- Understanding the data structure
- Making API calls
- Flattening the json output
About the open FDA project
There is so much to be discovered in publicly available datasets, for example, the US FDA has a large set of curated database that they have kindly made available for all to use, alongside with a nicely developed API. There are currently 6 main endpoints, ranging from information regarding animal and veterinary products, prescription and OTC drugs, to health devices and other information as the database gets updated (learn more from their website). To explore that, I’d like to start with their drug events endpoint, which gathers adverse events for prescription and over the counter medicines submitted voluntarily to the FDA.
Some prerequisites:
Below are some packages needed for the code:
import requests
import os
# import personal API key
import constant
import pandas as pd
import json
from pandas.io.json import json_normalize
import numpy as np
import time
import datetime
from joblib import delayed, Parallel
from ruamel.yaml import YAML
from pathlib import Path
# read in list of searchable fields
path = Path('fields.yaml')
yaml = YAML(typ='safe')
fields = yaml.load(path)
keys = constant.keys
pd.options.display.max_columns = None
Note:
I’ve downloaded the metadata yaml file for this endpoint to better understand the data structure, it is imported in the following code as
fields.yaml. More information can be found on thesearchable fieldssession on their website.I’ve also requested an API key (free) which is stored in a file called
constant.py
Understanding the data structure:
The json output of this API is highly nested, which can be a pain to deal with. Fortunately, with the metadata we can find out just which keys represent nested arrays, so we can work on flatten them later. To do that, I’ve written a script to parse through the YAML file.
Inspecting the searchable fields
The metadata fields is loaded in as a dictionary, specifically, properties is the attribute we’d like to dive into. Using keys() will reveal the list of keys within properties. The function get_key_type() below shows one way to parse through the list of keys and identify which one is an array or object:
def get_key_type(fields):
try:
field_key_list = [s for s in fields.get('properties',[]).keys()]
except:
raise Exception('Invalid input')
field_key_list = []
if field_key_list:
field_prop = [fields.get('properties',[]).get(s,'None') for s in field_key_list]
field_types = [field_prop[i]['type'] for i in range(0,len(field_prop))]
#print(field_types)
field_objs = [i for i in range(len(field_types)) if (field_types[i] == 'array') or (field_types[i] == 'object')]
print('The following keys are arrays or objects:')
print([field_key_list[i] for i in field_objs])
#print([field_prop[i] for i in field_objs])
return {'field_key_list':field_key_list,'nested_keys':field_objs,'nested_key_names':[field_key_list[i] for i in field_objs]}
else:
return None
Applying get_key_type() to fields, we’ll get a list of nested arrays. For example, we can tell that patient is a nested key, and running get_key_type() again on fields['properties']['patient'] reveal that within patient there are more nested arrays as well, which are: ['drug', 'patientdeath', 'reaction', 'summary']. We can then use this to further parse through the nested keys and find out the elements within each layer:
class get_fda_keys:
def __init__(self,fields):
self.key_types = get_key_type(fields=fields)
field_key_list = self.key_types['field_key_list']
nested_keys = self.key_types['nested_key_names']
# nested level = 0
self.single_keys = [i for i in set(field_key_list) - set(nested_keys)]
# nested level = 1
self.patient_keys = [val for val in set(fields['properties']['patient']['properties'].keys())- {'drug','reaction'}]
self.reaction_keys = [val for val in set(fields['properties']['patient']['properties']['reaction']['items']['properties'].keys())]
# nested level = 2
self.drug_keys = [val for val in set(fields['properties']['patient']['properties']['drug']['items']['properties'].keys()) - {'openfda'}]
# nested level = 3
self.openfda_keys = [val for val in set(fields['properties']['patient']['properties']['drug']['items']['properties']['openfda']['properties'].keys())]
self.primsource_keys = [val for val in set(fields['properties']['primarysource']['properties'].keys()) - {'literaturereference'}]
The class above will put all the nest keys together into an object for later use. Calling this:
fda_keys = get_fda_keys(fields= fields)
will print the following and return the list of other key names:
The following keys are arrays or objects:
['patient', 'primarysource', 'receiver', 'reportduplicate', 'sender']
List of variables from one of the nested keys:
For example, within the patient attribute is multiple sets of nested arrays, one of them is called drugs and we can parse through the structure using the following code:
self.drug_keys = [val for val in set(fields['properties']['patient']['properties']['drug']['items']['properties'].keys()) - {'openfda'}]
will return a list of column names within the drug key nested inside patient (notice that, I’ve removed openfda as that is another nested key):
Show list of drug related variables here:
['medicinalproduct',
'drugenddate',
'drugstructuredosageunit',
'drugtreatmentdurationunit',
'drugindication',
'drugadditional',
'drugdosageform',
'drugseparatedosagenumb',
'drugstructuredosagenumb',
'drugstartdateformat',
'drugrecurreadministration',
'drugbatchnumb',
'drugenddateformat',
'drugauthorizationnumb',
'drugcumulativedosagenumb',
'drugadministrationroute',
'drugcumulativedosageunit',
'drugintervaldosagedefinition',
'drugdosagetext',
'actiondrug',
'drugstartdate',
'drugcharacterization',
'drugintervaldosageunitnumb',
'activesubstance',
'drugtreatmentduration',
'drugrecurrence']
Making API calls
Now that we got a sense of the data structure, we can make some API calls using the requests package. To make a call, all we need is a url following the anatomy of the API endpoint, and an API key which can be obtained for free.
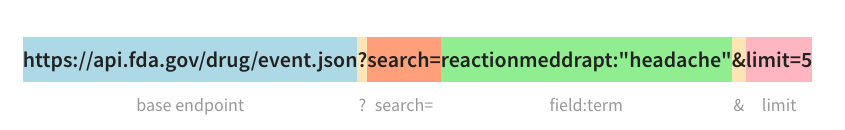
Using the following function, we can query the API using a range of receivedate as a parameter:
## API request:
def fda_api(key,sd,ed):
time.sleep(1)
# initialize output dataframe
output = pd.DataFrame()
base_url = 'https://api.fda.gov/drug/event.json?api_key='+key
search_term = '&search=receivedate:['+str(sd)+'+TO+'+str(ed)+']+AND+patient.patientonsetage:[1.0+TO+40000.0]+AND+patient.patientsex:[1+TO+2]&sort=receivedate:desc&limit=100'
url = base_url + search_term
r = requests.get(url)
if r.status_code == 200:
print('query succesful, getting data now:')
data = r.json()
return data
else:
print('Query unsuccesful, status_code: '+str(r.status_code))
return None
The function returns a dictionary, and from the metadata we can tell that the data is stored in the results section of it.
test = fda_api(key=keys['API_KEY'],sd ='20200101',ed = '20200102')
test.get('results')[0].keys()
which indicates the following attributes. From the exercise above, we already know a few of them are nested keys, which we’ll need to flatten, assuming we want to work with a dataframe later on:
dict_keys(['receiptdateformat', 'receiver', 'seriousnessdeath', 'companynumb', 'receivedateformat', 'primarysource', 'transmissiondateformat', 'fulfillexpeditecriteria', 'safetyreportid', 'sender', 'receivedate', 'patient', 'seriousnesshospitalization', 'transmissiondate', 'serious', 'receiptdate'])
Flattening the json output
One ofthe main nested attribute from the json output is the patient key, within it we can also find drug and openfda like so:
- patient
- drug
- openfda
- drug
To flatten that, we can use list comprehension to parse out each element of the nested key, for example:
for key in fda_keys.patient_keys:
patient_val = [json_out2[i]['patient'].get(key) for i in range(0,len(json_out2))]
df_list['patient_pdf'][key] = patient_val
print('df from nested patient keys')
print( df_list['patient_pdf'].shape)
The idea here is that we’ll then populate the collection of data frames which is defined by df_list. Once we have everything, we can then stitch them back together using pd.concat().
See the code to create df_list
#get keys from metdata:
fda_keys = get_fda_keys(fields = fields)
# get names for the keys:
df_names = [s.replace('keys','') for s,v in fda_keys.__dict__.items() if 'keys' in s]
# initiate empty data frames for each (nested) key:
df_list = dict((name+'pdf', pd.DataFrame()) for name in df_names)
There are probably far better ways to flatten the json structure than the following way, which is clumsy and not very pythonic, but for the time being it does what I need it to, which is parse through each of the nested keys and get the information it contains.
Let’s see the whole thing!
def get_pdf(sd,ed,fields = fields, keys = keys):
print('sd: ' + sd)
print('ed: ' + ed)
# API request:
json_out = fda_api(keys['API_KEY'],sd = sd,ed = ed)
json_out2 = json_out.get('results',[])
# get keys from metdata:
fda_keys = get_fda_keys(fields = fields)
# get names for the keys:
df_names = [s.replace('keys','') for s,v in fda_keys.__dict__.items() if 'keys' in s]
# initiate empty data frames for each (nested) key:
df_list = dict((name+'pdf', pd.DataFrame()) for name in df_names)
try:
for key in fda_keys.single_keys:
value = [json_out2[i].get(key,'None') for i in range(0,len(json_out2))]
df_list['single_pdf'][key] = value
print('df from unested keys')
print(df_list['single_pdf'].shape)
for key in fda_keys.primsource_keys:
primsource_val = [json_out2[i]['primarysource'].get(key,'None') if json_out2[i]['primarysource'] is not None else 'None' for i in range(0,len(json_out2))]
df_list['primsource_pdf'][key] = primsource_val
for key in fda_keys.patient_keys:
patient_val = [json_out2[i]['patient'].get(key) for i in range(0,len(json_out2))]
df_list['patient_pdf'][key] = patient_val
print('df from nested patient keys')
print( df_list['patient_pdf'].shape)
# some hard coded stuff until I fully figured out the data structure
case_event_date = [df_list['patient_pdf']['summary'][i].get('narrativeincludeclinical','None') if df_list['patient_pdf']['summary'][i] is not None else 'None' for i in range(0,len(df_list['patient_pdf'])) ]
df_list['patient_pdf']['case_event_date'] = case_event_date
for key in fda_keys.reaction_keys:
reaction_val = [json_out2[i]['patient']['reaction'][0].get(key,'None') for i in range(0, len(json_out2))]
df_list['reaction_pdf'][key] = reaction_val
for key in fda_keys.drug_keys:
drug_val = [json_out2[i]['patient']['drug'][0].get(key) for i in range(0,len(json_out2)) ]
df_list['drug_pdf'][key] = drug_val
active_substance = [df_list['drug_pdf']['activesubstance'][i].get('activesubstancename','None') if df_list['drug_pdf']['activesubstance'][i] is not None else 'None' for i in range(0,len(df_list['drug_pdf'])) ]
df_list['drug_pdf']['active_substance'] = active_substance
print('df from nested drug keys')
print(df_list['drug_pdf'].shape)
for key in fda_keys.openfda_keys:
openfda_val = [get_openfda_ind(json_out2[i],key) for i in range(0,len(json_out2))]
#print(openfda_val)
df_list['openfda_pdf'][key] = openfda_val#[openfda_keys.index(key)]
print('df from nested openfda keys')
print(df_list['openfda_pdf'].shape)
total_pdf = pd.concat([df_list[name] for name in df_list],axis = 1)
total_pdf['patientonsetageunit'] = total_pdf['patientonsetageunit'].apply(lambda x : float(x) if x is not None else np.NaN)
total_pdf['patientonsetage'] = total_pdf['patientonsetage'].apply(lambda x : float(x) if x is not None else np.NaN)
real_age = [age_convert(total_pdf['patientonsetage'][i],total_pdf['patientonsetageunit'][i]) for i in range(0,len(total_pdf))]
total_pdf['patient_age_year'] = real_age
serious_flags = [val for val in total_pdf.columns if 'seriousness' in val]
print(serious_flags)
for flag in serious_flags +['reactionoutcome']:
total_pdf[flag] = total_pdf[flag].apply(lambda x : 0 if x == 'None' else 1)
print('resulting data dimension:')
print(total_pdf.shape)
return total_pdf
except:
return None
Testing out the function:
test_df = get_pdf(sd = '20200101',ed = '20200102')
test_df.head(3)
The result is a pretty large list of variables since we have flatten all the nested keys from the initial json output.
| companynumb | receivedate | seriousnessdisabling | seriousnesshospitalization | seriousnesscongenitalanomali | seriousnesslifethreatening | authoritynumb | safetyreportid | safetyreportversion | transmissiondate | ... | rxcui | unii | manufacturer_name | product_type | pharm_class_epc | route | spl_set_id | pharm_class_moa | reportercountry | qualification | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | US-004585 | 20110101 | None | None | None | None | None | 7288847-8 | None | 20110831 | ... | ['2170286', '238720', '883826', '208104'] | ['6T84R30KC1'] | ['Amneal Pharmaceuticals LLC', 'Akorn, Inc.', ... | ['HUMAN OTC DRUG', 'HUMAN PRESCRIPTION DRUG'] | None | ['TOPICAL', 'ORAL', 'INTRAVENOUS'] | ['6326bdbf-fb2e-451a-a0a0-7d0b59ce5438', '031b... | None | UNITED STATES | 1 |
| 1 | US-PURDUE-USA-2010-0060882 | 20110101 | None | None | None | None | None | 7205909-1 | None | 20110831 | ... | ['1860137', '1860157', '1860129'] | ['C1ENJ2TE6C'] | ['KVK-Tech, Inc.'] | ['HUMAN PRESCRIPTION DRUG'] | None | ['ORAL'] | ['c5b52ff1-21a6-4d28-9982-55b4ac195fac'] | None | UNITED STATES | 5 |
| 2 | 813209 | 20110101 | None | None | None | None | None | 7288697-2 | None | 20110831 | ... | ['846192', '197577', '309686', '309684', '8541... | ['7S5I7G3JQL'] | ['Gentex Pharma', 'Allergan, Inc.', 'West-Ward... | ['HUMAN PRESCRIPTION DRUG'] | None | ['INTRACANALICULAR', 'INTRAOCULAR', 'ORAL', 'I... | ['f8d52954-be1d-45d3-9245-ff2e3e81a8ac', 'b152... | None | AUSTRALIA | 3 |
| 3 | ES-ELI_LILLY_AND_COMPANY-ES201012004665 | 20110101 | None | None | None | None | None | 7205908-X | None | 20110831 | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | SPAIN | 5 |
4 rows × 80 columns